There are three sections of this website that provide a search function for toxins:
(1) Go to "Search for Toxin" from the search drop-down menu on the homepage;
(2) Go through the search bar in the "Search for BioTD" section on the homepage;
(3) Go through the "Toxin Search" button of the home page of "Function in BioTD";
In the field of “Search for BioTD”, users can find toxin entries by searching toxin name, target name, and so on among the entire textual component of BioTD. Query can be submitted by entering keywords into the main searching frame. The resulting webpage displays profiles of all the toxins directly related to the search term, including Toxin name, Toxin species, Representative target, Alternative name, Target family , Target species and their information links.
In order to facilitate a more customized input query, the wild characters of “*” and “?” are also supported.
(1) If search “Apamin”, find a single API entry which is named “Apamin”;
(2) If search: “Shk”, finds a series entries with toxin names;
(3) If search: “Venom plasminogen activator Haly*”, find a single entry which is named “Venom plasminogen activator Haly-PA”;
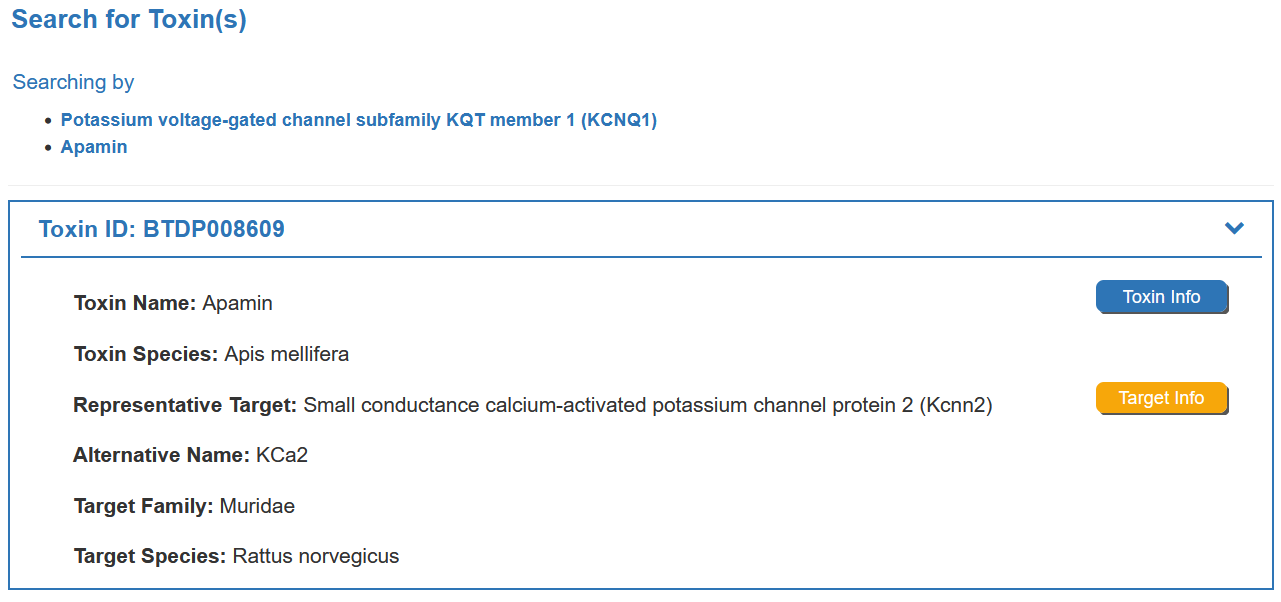
For example, if you want to know the detail information for Apamin, you can search Apamin in the “Search for Toxin” field.
Search result of the “Apamin” shows the information of Toxin ID, Toxin Name, Representative Target, Representative Target, Target Family, Target Species and External buttons. The “Toxin Info” button links to the detailed toxin information page of Apamin. The “Target Info” button links to the detailed target information page of Apamin.
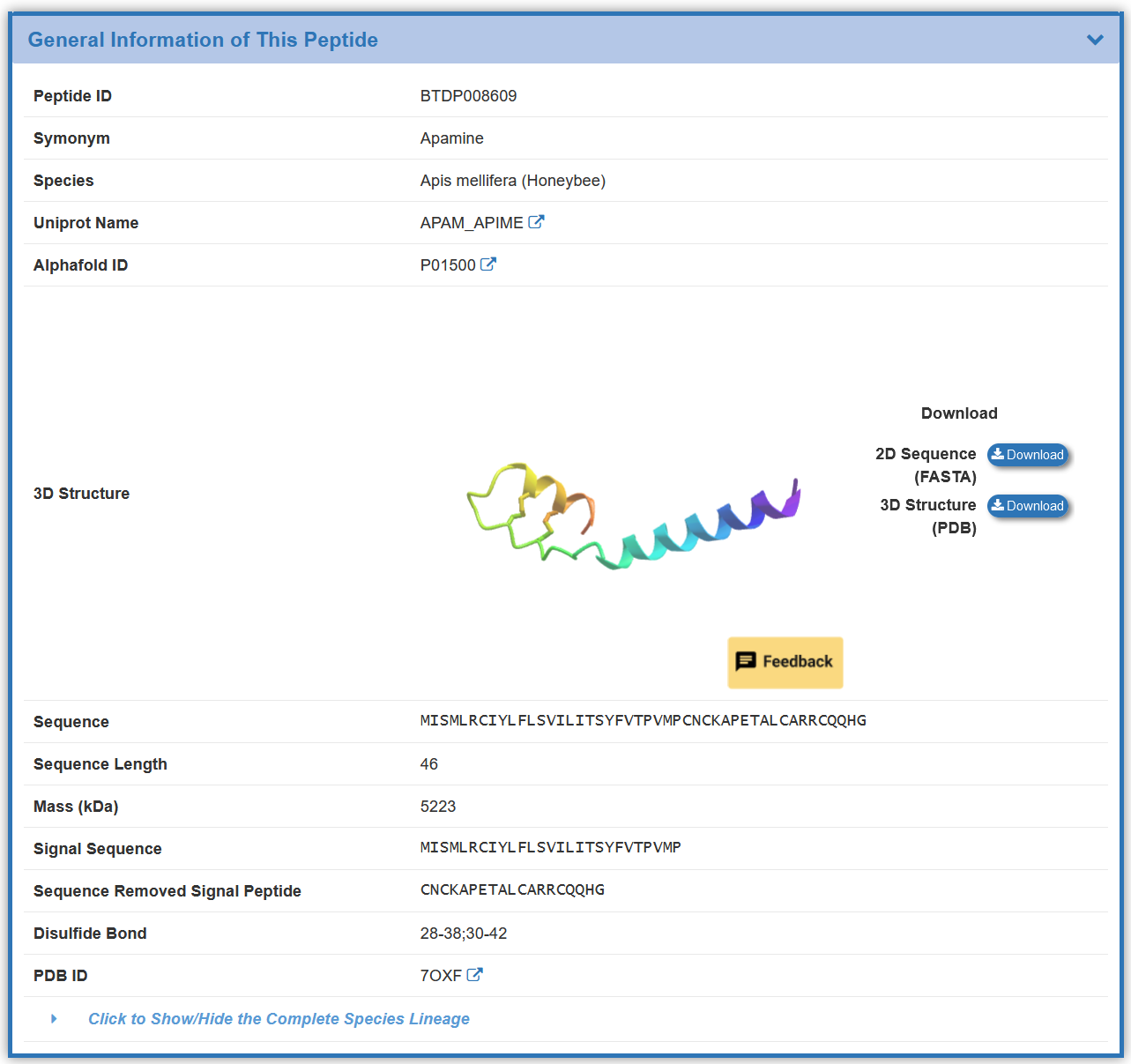
1.1. By clicking the “Toxin Info” button, the detailed information page for particular toxin will be displayed
Take 'Potassium channel toxin alpha-KTx 3.6' as an example, “General Information of This Peptide” section displays the general information of Psalmotoxin-1 including its ID, Name, Symonym, Species, Uniprot Name, Alphafold ID, 3D Structure, Sequence, Sequence Length, Mass (kDa) ,Sequence Removed Signal Peptide, Disulfide Bond, PDB ID ,Species-lineage.
In the 3D Structure section, the peptide structures are sourced from the PDB, the AlphaFold Database, or predicted using ColabFold v1.5.5 (hover the mouse to view AlphaFold2 parameters).
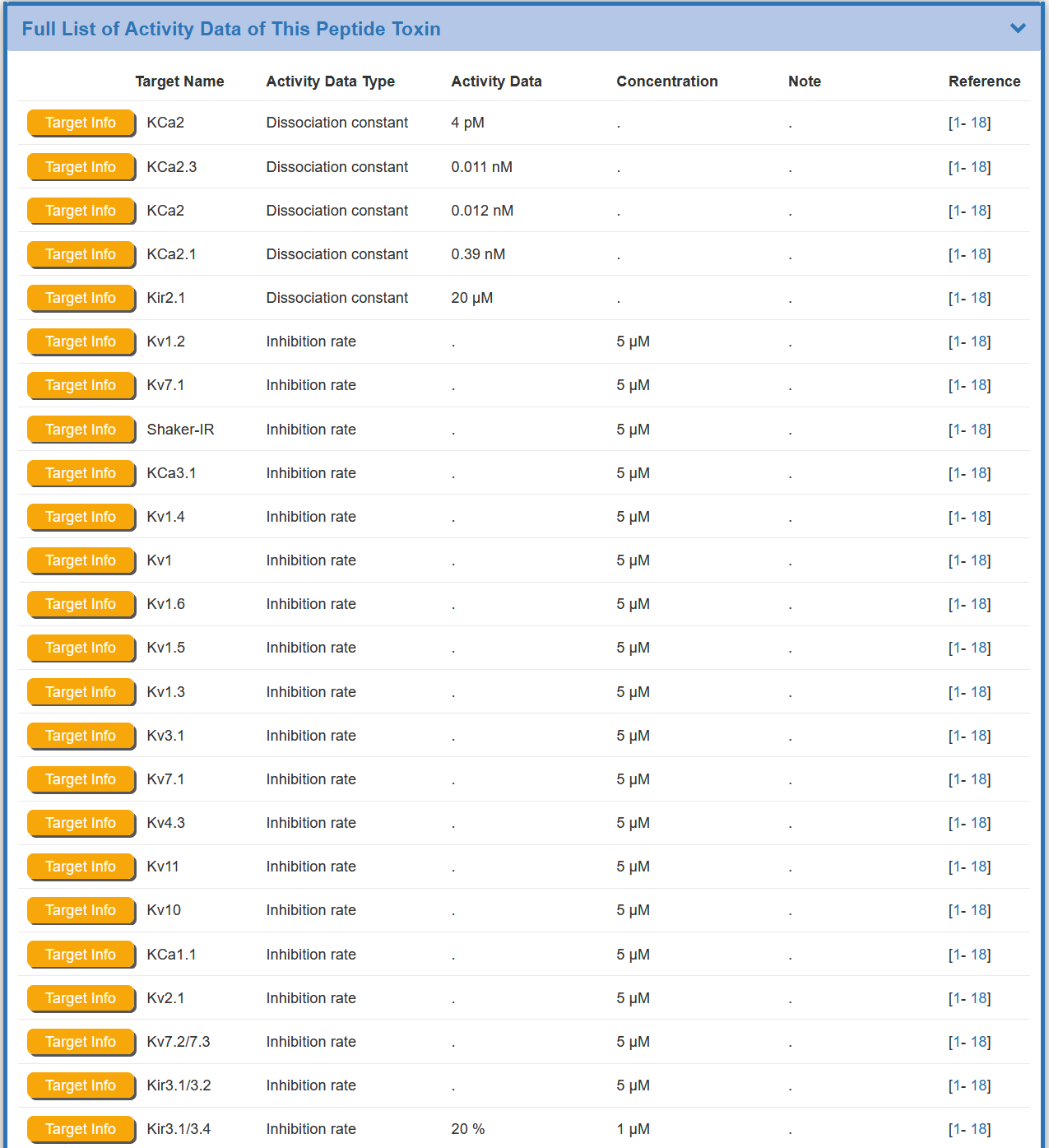
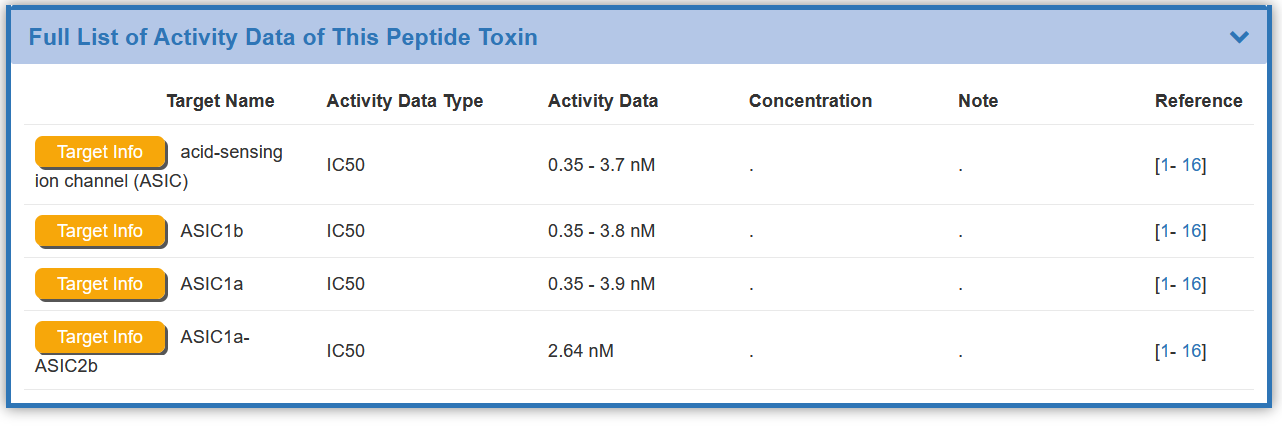
Some of the toxin(s) in the BioTD have corresponding target(s), and the activity of most targets has been determined by various experimental pathways The “Full List of Activity Data of This Peptide Toxin” section shows the Target Name, Activity Data Type , Activity Data ,Concentration , Note ,and Reference.
1.2. By clicking the “Target Info” button, the detailed target information page will be displayed
Take Kcnn2 as an example, “General Information of This Target” section displays the general information of target, including its ID, Name, Bioclass, Uniprot ID, 3D Structure, Gene Name, Gene ID, Synonym, Sequence, Family, Function, Taxonomy ID and External Link(s).
In the 3D Structure section, the target structures are sourced from the PDB, the AlphaFold Database, or predicted using ColabFold v1.5.5 (hover the mouse to view AlphaFold2 parameters).
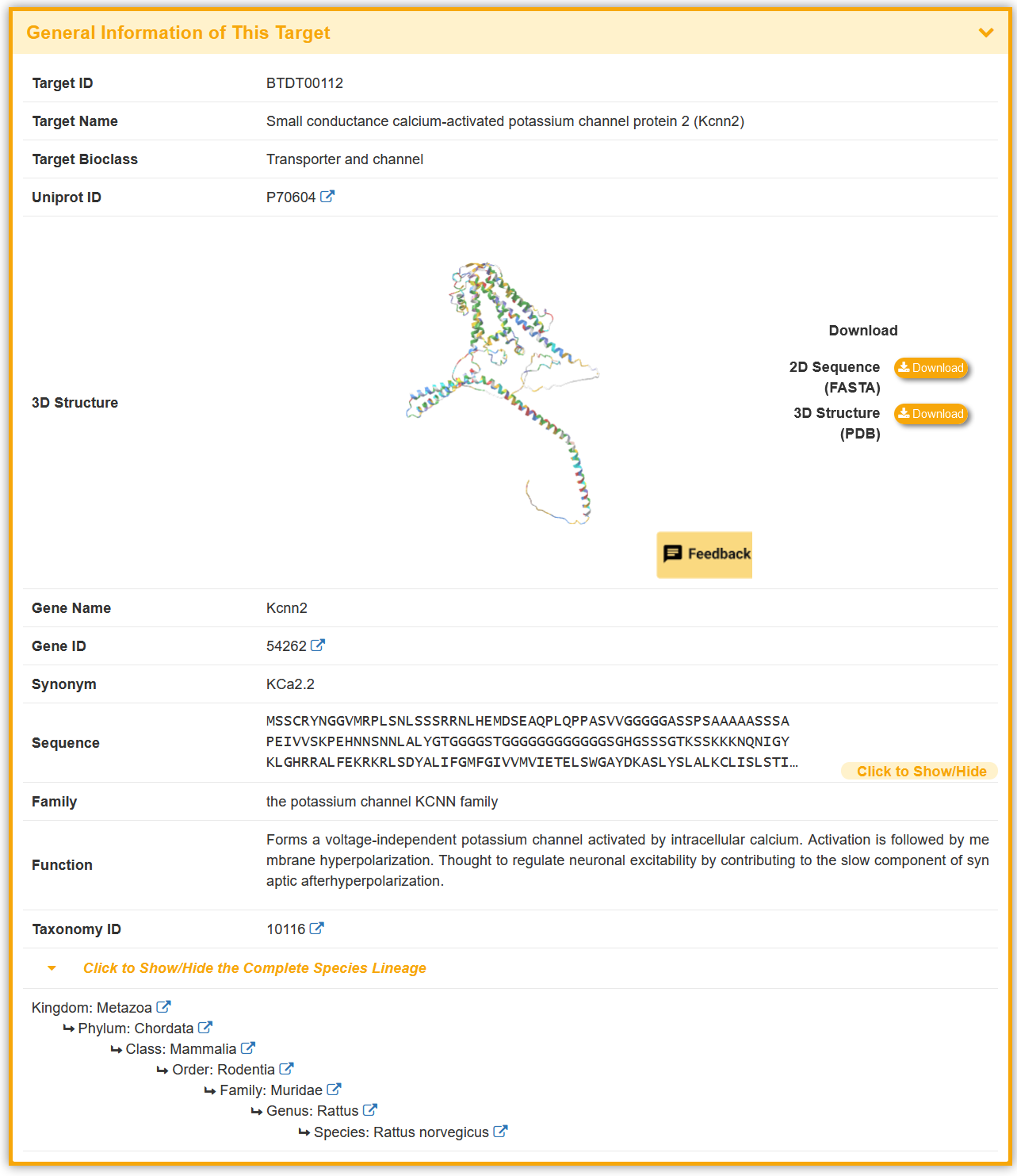
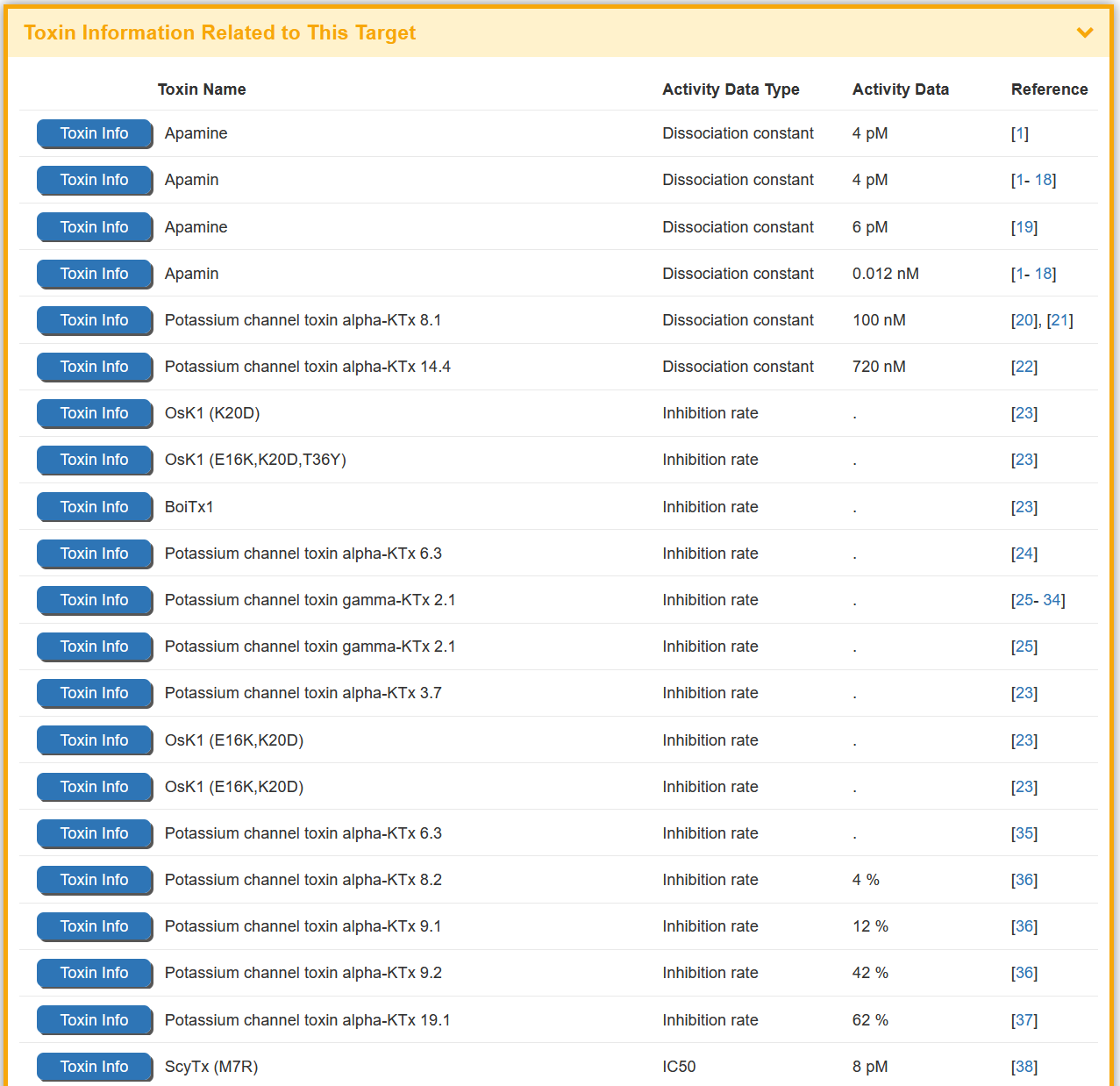
Some of the toxin(s) in the BioTD have corresponding binding activity, and are evaluated using various index. The “Toxin Information Related to This Target” section shows the Toxin Name, Activity Data Type, Activity Data,and Reference.
There are two sections of this website that provide a search function for targets.
(1) Go to "Search for Target" from the search drop-down menu on the homepage;
(2) Go through the "Target Search" button of the home page of "Function in BioTD";


In the field of “Search for Target”, users can find target entries by searching target ID, target name, toxin name, gene name and so on among the entire textual component of BioTD. Query can be submitted by entering keywords into the main searching frame. The resulting webpage displays profiles of all the targets directly related to the search term, including Target name, Alternative name, Target bioclass, Target species, Representative toxin ,Toxin species, and their information links.
In order to facilitate a more customized input query, the wild characters of “*” and “?” are also supported.
(1) If search “Nav1.7”, find two API entries which is named “Nav1.7”;
(2) If search: “KvA*”, find a single entry which is named “KvAP”;
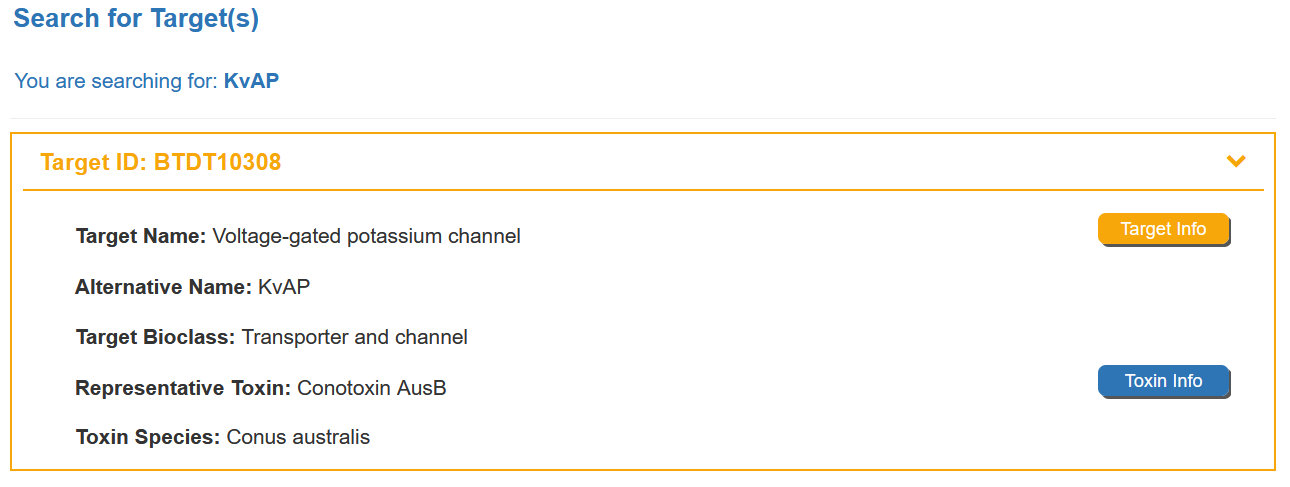
For example, if you want to know the detail information for KvAP, you can search “KvAP” in the “Search for Target” field.
Search result of the “KvAP” shows the information of Target ID, Target Name, Alternative Name, Target Bioclass, Representative Toxin ,Toxin Species and External buttons. The “Target Info” button links to the detailed target information page of Voltage-gated potassium channel. The “Toxin Info” button links to the detailed toxin information page of Conotoxin AusB.
2.1. By clicking the “Target Info” button, the detailed target information page will be displayed
Take ASIC1a as an example, “General Information of This Target” section displays the general information of target, including its ID,Name, Synonyms, Bioclass, Uniprot ID, 3D Structure, Gene Name, Gene ID, Synonym, Sequence, Family, Function, Taxonomy ID and External Link(s).
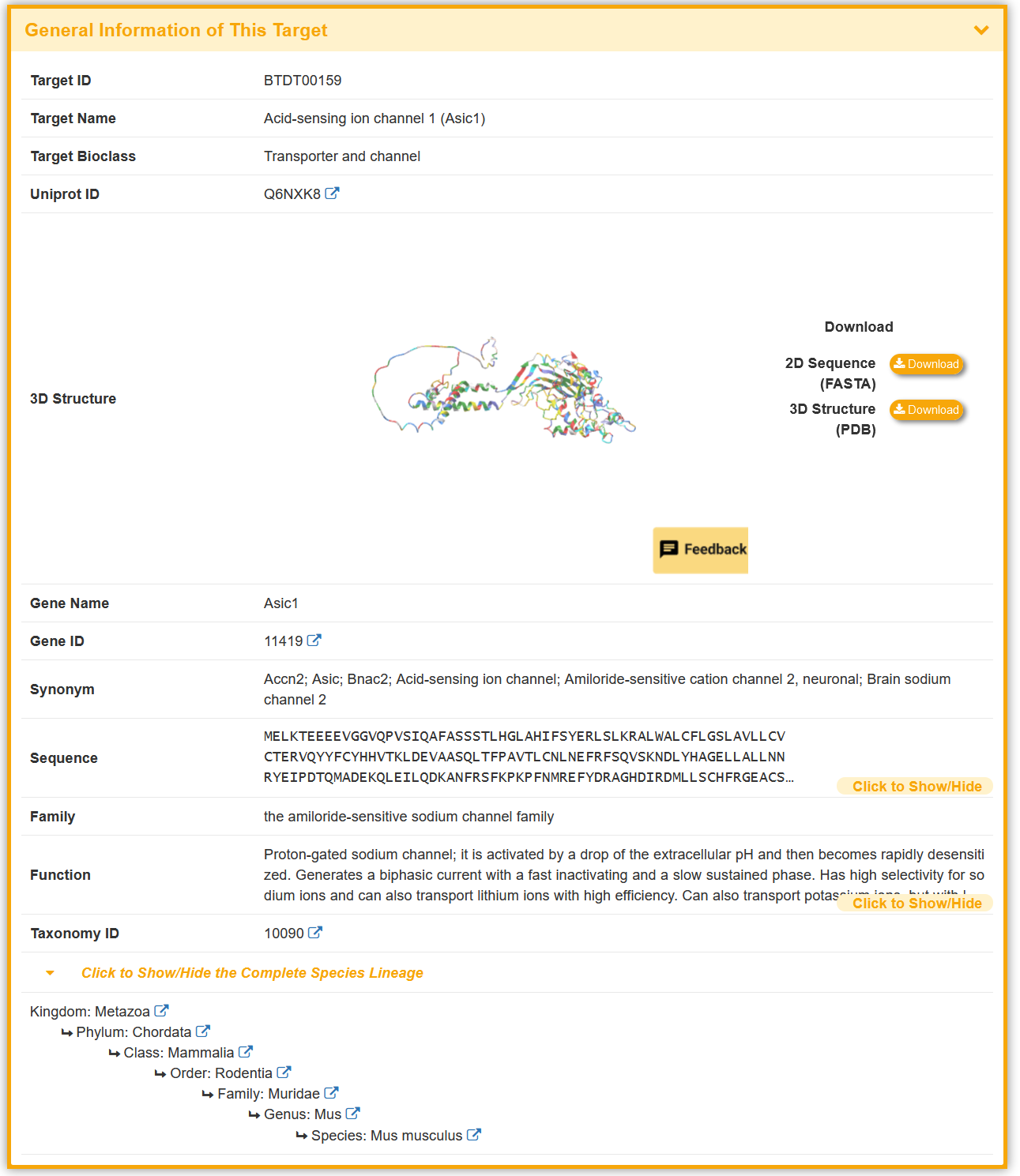
Some of the targets in the BioTD have corresponding binding activity, and are evaluated using various index. The “Toxin Information Related to This Target” section shows the Toxin Name, Activity Data Type, Activity Data and Reference.
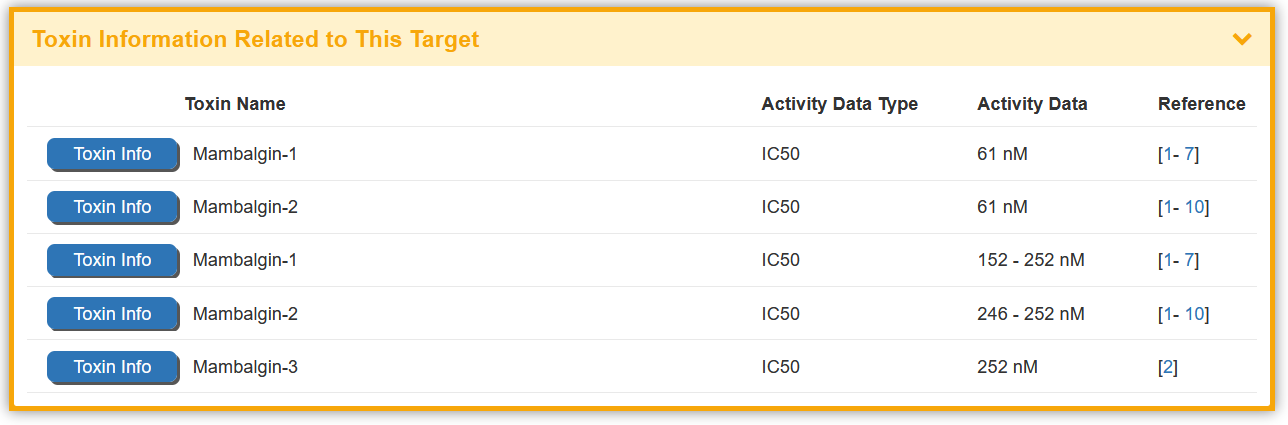
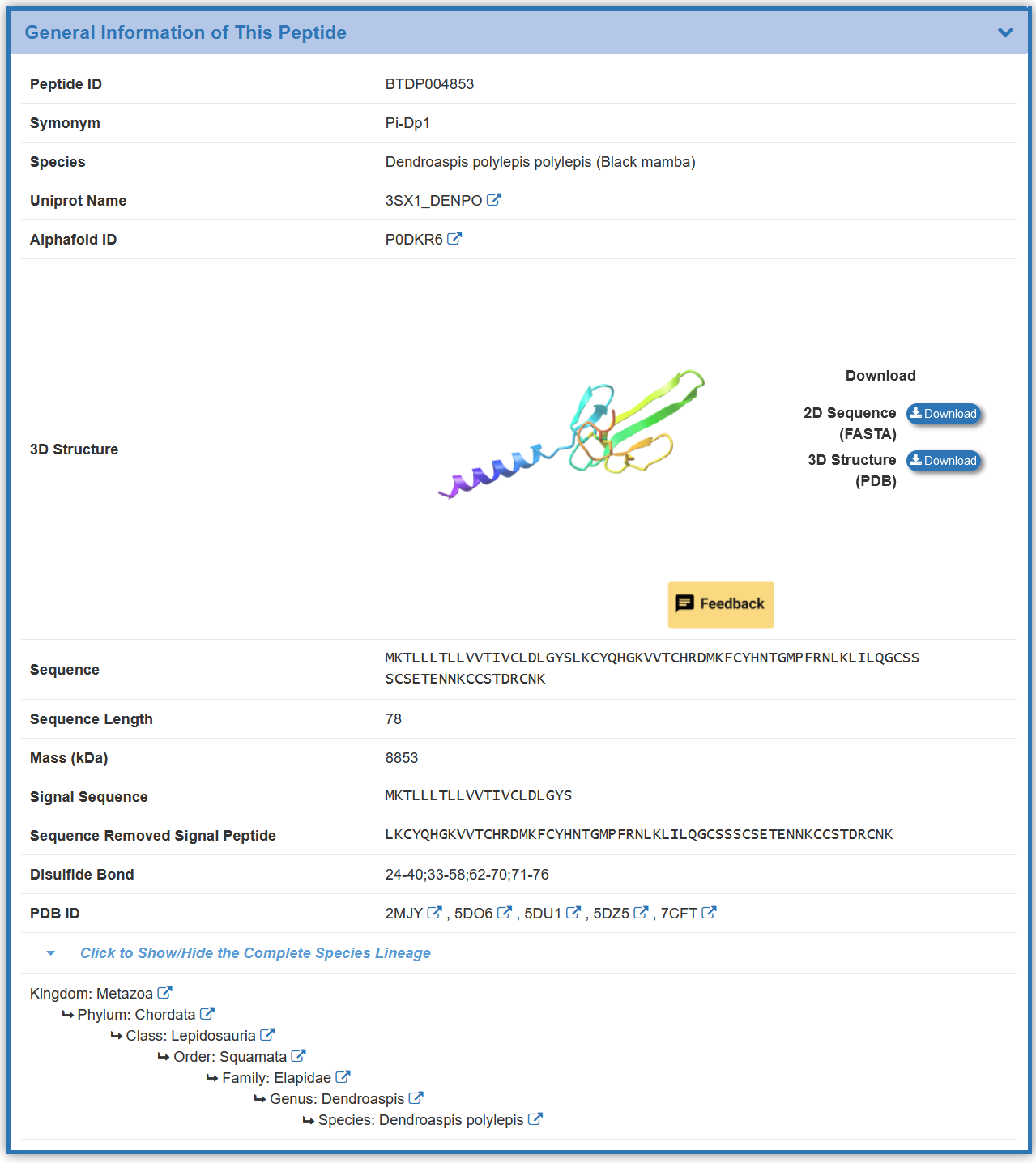
2.2. By clicking the “Toxin Info” button, the detailed information page for particular toxin will be displayed
Take Psalmotoxin-1 as an example, “General Information of This Peptide” section displays the general information of Psalmotoxin-1 including its ID, Name, Symonym, Species, Uniprot Name, Alphafold ID, 3D Structure, Sequence, Sequence Length, Mass (kDa) ,Sequence Removed Signal Peptide, Disulfide Bond, PDB ID ,Species-lineage.
Some of the toxin(s) in the BioTD have corresponding binding activity, and are evaluated using various index. The “Toxin Information Related to This Target” section shows the Toxin Name, Activity Data Type, Activity Data,and Reference.
There are two sections of this website that provide a search function for sequence similarity search in BioTD.
(1) Go to "Sequence Similarity Search" from the search drop-down menu on the homepage;
(2) Go through the "Toxin Sequence Similarity Search" button of the home page of "Function in BioTD";


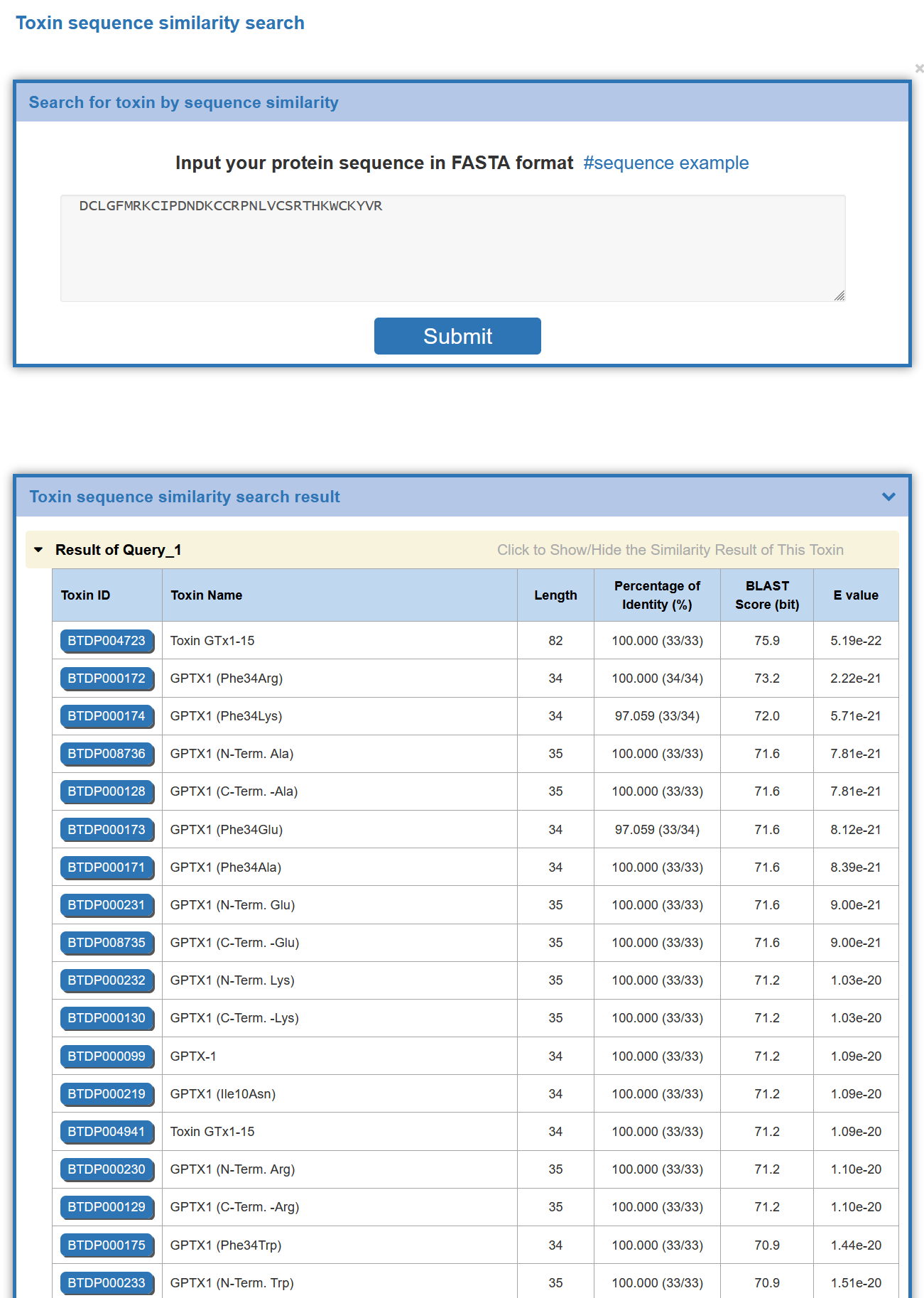
In the field of “Sequence Similarity Search”, a BLASTp-based molecular sequence similarity search method is also provided in BioTD. Users can access it through http://biotoxin.net/search/sequence-similarity Toxin sequence similarity search.

On this page, the user can enter and submit the sequence in FASTA format of any protein, and BioTD will perform a sequence-based protein similarity search.This page provides a sequence of examples.
The similarity degree of identified molecules will be evaluated by the BLAST program, and with the BLAST E-value listed out in the order from the highest to the lowest. Links to the detailed information of identified toxins are also provided.
On the search results page, BioTD provides a list of toxins with significant similarity to the sequence entered by the user, all molecules are sorted by E-value size, and the results table provides " Toxin ID","Toxin Name", "Length", "Percentage of Identity (%)", "BLAST Score (bit)", "E-value", etc. The user can click on the Toxin ID to access the toxin details page.